Visium Spatial Genomics: Discontinued.
Visium spatial gene expression measures total mRNA with morphological context in intact tissue samples, both fresh frozen or FFPE. This can be used to map whole transcriptomes in order to better investigate disease pathology, development, and clinical translational research.
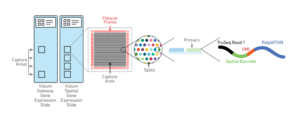
Spatial gene expression slides contain 4 capture areas (6.5 x 6.5mm) bordered by a fiducial frame (8 x 8 mm with the frame). Each capture area contains ~5,000 gene expression “spots”, which in turn contains primers comprised of the following:
- Illumina TruSeq Read 1
- 16 nt Spatial Barcode
- 12 nt unique molecular identifier (UMI)
- 30 nt poly(dt) sequence (to capture poly-adenylated mRNA for cDNA synthesis)
Consultation and Assistance
Contact for Genomics and Bioinformatics Consultation
Details about sample preparation, library prep types, and application are all listed on this page. For more information and assistance contact ggbc@uga.edu.
Workflow
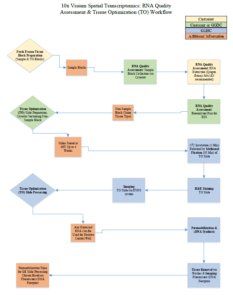
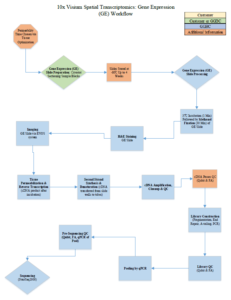
10x Visium Spatial Transcriptomics Workflow
| Step in Workflow | Summary/Notes | 10x Guide and Link | Customer or GGBC | 10x How-To Videos |
|---|---|---|---|---|
| Fresh Frozen Tissue Block Preparation | Tissue block preparation; any block of the same tissue type can be used for TO step | Link to Tissue Prep Guide (refer to pg. 5-9) | Completed by customer; protocols for simultaneous or separate embedding/freezing | Link to Tissue Preparation Guide Video |
| RNA Quality Assessment Sample Collection | Tissue collection from all sample blocks for RNA extraction | Link to Tissue Prep Guide (refer to pg. 17-18) | Either customer uses cryostat (hourly rate) or GGBC collects with customer present | |
| RNA Quality Assessment RNA Extraction | RNA extraction using Qiagen RNeasy Mini Kit | Link to Tissue Prep Guide (refer to pg. 17-18) | Completed by customer | |
| RNA Quality AssessmentBioanalyzer Run | Run RNA on bioanalyzer for RIN scores | Link to Tissue Prep Guide (refer to pg. 17-18) | Either by customer or GGBC bioanalyzer order | |
| Tissue Sectioning/Slide Preparation (TO & GE) | Tissue permeability time optimization | Link to Tissue Prep Guide (refer to pg. 10-16) | Either customer uses cryostat (hourly rate) or GGBC sections & assembles slide | |
| Methanol Fixation, H&E Staining (TO & GE) | Tissue fixation & staining | Link to Fixation & Staining Protocols | Completed by GGBC | |
| Imaging (TO & GE) | EVOS system | Link to Imaging Guidelines | Completed by GGBC | |
| Tissue Optimization (TO) Slide Processing | Determination of permeability time | Link to Tissue Optimization User Guide | Completed by GGBC | Link to TO Protocol Video |
| Gene Expression (GE) Slide Processing | Permeability Step through Library Construction | Link to Gene Expression User Guide | Completed by GGBC | Link to GE Protocol Video |
| Pre-Seq QC & Sequencing | Link to Gene Expression User Guide (pg. 59-61) | Completed by GGBC |
TO = Tissue Optimization GE = Gene Expression
Helpful Links Related to Visium Spatial Genomics Sample Preparation
1. Tissue Permeability Optimization
https://assets.ctfassets.net/an68im79xiti/3jBtg0tFMD28GaEqoStahT/2cc85436f97d5b9192e8201cf4961d88/CG000238_VisiumSpatialTissueOptimizationUserGuide_RevE.pdf
For new projects / new tissue types, tissue permeability optimization will need to be performed. This process ensures the optimal permeabilization time is chosen; this time point should result in maximum fluorescence signal with the lowest signal diffusion. Each tissue type will require its own permeability optimization. Tissue permeability does NOT need to be performed on the specific sample block, but rather the same tissue TYPE. Please clearly label the block(s) and indicate on the order form which block(s) should be utilized for either Gene expression or tissue optimization.
The cryostat at the core can be utilized by researchers for a small fee. Please contact GGBC (ggbc@uga.edu) to schedule this service. The customer may prepare both their tissue optimization slide and gene expression slide simultaneously. The tissue optimization slide will be stored in the -80°C until tissue optimization is complete. Both slides are stable in the -80°C for up to 4 weeks.
2. Slide Preparation
Tissue Sectioning / Slide Submission
Tissue sectioning can be done by the researcher, or fresh frozen / FFPE blocks can be provided to the core for sectioning.
Slides are stable for up to four weeks in -80°C and can be shipped on dry ice to the GGBC facility.
Gene Expression Slide Specifications
| Capture Area Type | Size | Barcoded Spots | Spot Size |
|---|---|---|---|
| Standard | 6.5mm x 6.5 mm | ~5000 | 55um |
| XL capture | 11 mm x 11 mm | ~14,000 | 55 um |
*1-10 cells will cover a spot, depending on the type of tissue used.
H&E Staining and Imaging
Slides are stained using hematoxylin and eosin (H&E) staining and are imaged using EVOS.
3.Permeabilize Gene Expression Tissue and Construct Library
Prices and Quotes
Contact for Financial Inquiries and Quote Requests
Please email Kim and Elizabeth at ggbc@uga.edu, for financial inquiries or to request a quote. Be as specific as possible, so that they can more quickly assist you.
A Note About Visium Spatial Transcriptomics Projects:
Please contact the core to inquire about pricing. Scheduling projects with other lab groups may be beneficial. If no other group(s) require tissue optimization around the time a customer needs this service, the core may charge the customer for the full price of the kit.
Visium Spatial Transcriptomics
| Step | Service | UGA Fee | Non-UGA Fee | Commercial Fee |
|---|---|---|---|---|
| 1. Tissue Permeability Optimization | One Slide | $1,383.00 | $1,632.00 | $1,729.00 |
| 4 Slides | $4,332.00 | $5,111.00 | $5,414.00 | |
| 2. Tissue Sectioning and Slide Preparation | Includes tissues sectioning, staining, and imaging a single slide with up to 4 capture areas. | $168.00 | $199.00 | $210.00 |
| 3. Tissue Gene Expression Library Prep | Includes cDNA synthesis, cDNA QC, library prep, library QC | $6,621.00 | $7,813.00 | $8,276.00 |
| 4. Library Pooling | Library Pooling | $60.00 | $71.00 | $75.00 |
| 5. Pre-Sequencing QC | QC the final library before preparing to load | $120.00 | $142.00 | $150.00 |
| 6. NextSeq 2000 Sequencing Run | Run can vary depending on output requirements. Typically P2 or P3. | Price will vary based on flow cell. | Price will vary based on flow cell. | Price will vary based on flow cell. |
| Cryostat Training | 1 hour training with designated laboratory personnel. After a customer is “trained” they are free to schedule cryostat use. | $50.00 | $59.00 | $63.00 |
| Cryostat Use | Trained customers are welcome to schedule cryostat usage in increments of 1 hour. | $15.00 | $18.00 | $19.00 |
Data Analysis
10X Space Ranger analysis software can be utilized to process spatial gene and/or protein expression data. This software can be utilized in conjunction with Loupe Browser visualization software.
Resources:
https://support.10xgenomics.com/spatial-gene-expression/software/pipelines/latest/what-is-space-ranger [Name “Space Ranger Analysis Pipelines”]
https://support.10xgenomics.com/spatial-gene-expression/software/visualization/latest/what-is-loupe-browser [Name “Loupe Browser Visualization software”]