Bioinformatics
Next-generation (NGS) and Third-Generation sequencing approaches are ubiquitous in biological, genetic and clinical experimentation. Researchers requiring manipulation and/or evaluation of these data often realize that either specialized guidance or higher-level bioinformatics expertise is needed to expand their capabilities and accomplish their goals.
The GGBC Bioinformatics team at UGA is comprised of experienced research faculty and graduate student interns, who apply best-practice methodologies and employ open-source and custom-built software for data processing, data analysis and visualization options for a wide range of NGS datasets.
Our consultation services are free for input on experimental design and proposal development. Some of the more common bioinformatics workflows, e.g. RNA-Seq, bacterial genome assembly, transcriptome assembly and variant analysis, are priced in an accessible, modular fashion (see Prices below). More complex analyses, e.g. eukaryotic genome assembly, comparative genomics and many customized workflows are priced using hourly rates, since it’s often tough to be accurate with estimates until the work starts.
Offerings
- Team consultation
- Experimental design
- Variety of computational and bioinformatics analyses
- Training on specific analysis upon request
- Customized analysis pipelines
Guidelines for Different Analysis Workflows
Microbiome 16S/18S/ITS Sequencing Data Analysis
- Type of Data: Targeted sequencing of any of the 16S or 18S or ITS regions.
- Analysis methods:
Quality-based sequence trimming and removal of adapters and specific primer sequences, removal of chimeric sequences, joining the forward and reverse reads, representative sequences identification, taxonomy classification, and statistical analysis. - Deliverables:
Species Richness per sample, Species Relative Abundance among samples, Core Microbiomes, Alpha-diversity, Beta-diversity, Differential Analysis
RNA-Seq
- Type of Data: Illumina PE 50, 75, 100, etc… stranded libraries. Required: annotated (GFF3/GTF) genome or transcriptome.
- Analysis methods:
Data Quality Assessment (Raw and Trimmed): FastQC
Data Quality Trimming: Trimmomatic
Mapping: STAR/Bowtie2, depending on input dataset.
Expression Analysis: DESeq2/edgeR/RSEM, depending on reference and dataset. - Deliverables:
Quality assessment for all samples (raw and trimmed), PCA and/or BCV analysis of samples, MA plots, list of differentially expressed genes in Excel format including fold change, P values, FDR, normalized counts, etc… as well as ancillary files, e.g. read mapping metrics, BAM files.
Small RNA (sRNA) Analysis
- Type of Data: Illumina SE75 (CAP-miRSeq requirements: reference genome, miRbase species accession)
- Analysis methods:
Data Quality Assessment (Raw and Trimmed): FastQC
Data Trimming: CutAdapt
Expression Analysis: CAP-miRSeq Pipeline (Bowtie, Randfold, HTSEQ, MIRDEEP2) - Deliverables:
Quality assessment for all samples (raw and trimmed), trimmed read distributions, profile of all small RNAs present in each sample, prediction of novel miRNAs.
General expression results: Excel files with mature, raw, normalized and novel miRNA counts.List of differentially expressed miRNAs in Excel format including fold change, P values, FDR, normalized counts.
SNP Analysis
- Type of Data: Illumina PE75. Mapping reference required.
- Analysis methods:
Data Quality Assessment (Raw and Trimmed): FastQ Data Trimming: Trimmomatic
Mapping and mark duplicates: BWA, Picard
Variant Calling and filtering: GATK - Deliverables:
Quality assessment for all samples (raw and trimmed), trimmed read metrics, mapping and read duplication stats, SNP and Indel variant call files (VCF) for both raw and filtered datasets.
Bacterial Genome Assembly & Annotation (short read)
- Type of Data: Illumina PE150, PE300
- Analysis methods:
Data Quality Assessment (Raw and Trimmed): FastQC
Data Trimming: Trimmomatic
Assembly: SPAdes
Benchmarking: Quast, BUSCO, BlastN, Mauve
Automated Annotation & prophage discovery: RASTtk, PHASTER - Deliverables:
Assembly fasta file, Quality assessments for all samples (raw and trimmed, Quast summary metrics (plus/minus reference), BUSCO identification of core gene set, Mauve alignment to closest genome reference and ordering of contigs, BlastN (tabular output), RASTtk annotation (xls, gff, gbk, peptide.fa), PHASTER identification of prophage sequence(s).
Bacterial Genome Assembly & Annotation (long read)
- Type of Data: PacBio
- Analysis methods:
Error correction, assembly and contig polishing: Canu, BLASR, Arrow
Benchmarking: Quast, BUSCO. - Deliverables:
Assembly fasta file, Contig coverage plots, Quast summary metrics (plus/minus reference), BUSCO identification of core gene set, Mauve alignment to closest genome reference and ordering of contigs, BlastN (tabular output), RASTtk annotation (xls, gff, gbk), PHASTER identification of prophage sequence(s).
Eukaryotic Genome Assembly (de novo, long read)/Custom
- Type of Data: Illumina PE
- Analysis methods:
Data Quality Assessment (Raw and Trimmed): FastQC
Data Trimming: Trimmomatic
Assembly: Velvet/Soapdenovo2/ABySS (depending on dataset)
Benchmarking: Quast, BUSCO, BlastN - Deliverables:
Assembly fasta file, Quast summary metrics (plus/minus reference), BUSCO identification of core gene set, BlastN (tabular output).
Transcriptome Assembly (short read)
- Type of Data: PE75, PE150
- Analysis methods:
Data Quality Assessment (Raw and Trimmed): FastQC
Data Trimming: Trimmomatic
Assembly: Trinity
Benchmarking: Quast, BUSCO - Deliverables:
Assembly fasta file, Quast summary metrics (plus/minus reference), BUSCO identification of core gene set.
Transcriptome Assembly (long read)
- Type of Data: PacBio
- Analysis methods:
Error Correction, assembly and contig polishing: IsoSeq3
Transcript clustering: Minimap2, Cupcake (+ reference genome), Cogent (-/+ reference genome)
Benchmarking: Quast, BUSCO - Deliverables:
High quality and low quality fasta and fastq transcript files, locus collapsed and 5’ degradation filtered assembly fastas and gff file. Quast summary metrics (plus/minus reference), BUSCO identification of core gene set.
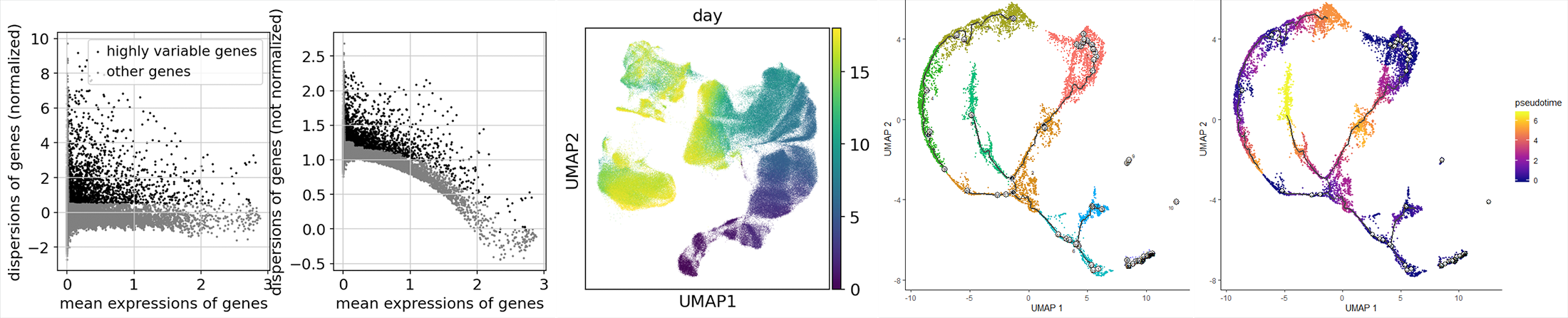
Single-cell RNA-Seq Data Analysis
- Type of Data: 10x Single-cell RNA-sequencing Data
- Analysis methods:
Data Quality Control
Data processing: Normalization, Principal component analysis (PCA) dimension reduction, Clustering/Community Detection.
Data visualization: t-Distributed Stochastic Neighbor Embedding (t-SNE)/ Uniform
Manifold Approximation and Projection (UMAP) cell visualization.
Cell Trajectories analysis, Pseudo-time analysis, Differential expression analysis, Single-cell time series analysis - Deliverables:
Comprehensive Report on all requested analysis including codes, figures and paper format writing explanations.
Analysis on CRISPR Genetic screens
- Type of Data: Genome-wide screen data
- Analysis methods:
Generating the raw count table using “MAGeCK” MAGeCK raw count and Batch effect remove. MAGeCK raw count and NO Batch remove. MAGeCK raw count and DEseq2 package analysis. - Deliverables:
Comprehensive Report on all requested analysis including codes, figures and paper format writing explanations.
Circular Consensus Sequencing (CCS) Read Generation
- Type of Data: PacBio Sequel II
- Analysis methods:
Generation of CCS reads resulting from alignment between subreads taken from a single ZMW. CCS reads are advantageous for amplicon, RNA, and genome sequencing projects and are highly accurate (>99% accuracy, Q>20). - Deliverables:
A CCS.bam file for each SMRT cell run and a text file containing read generation statistics and metrics.
Prices
| Service | UGA Fee | Non-UGA Fee | Commercial Fee |
|---|---|---|---|
| De novo transcriptome assembly from illumina short reads | $1,500 | $1,770 | $1,875 |
| De novo transcriptome assembly from PacBio Iso-Seq data | $1,500 | $1,770 | $1,875 |
| Assembly of small (< 50 Mb) eukaryotic genomes | custom | custom | custom |
| Microbial genome assembly & annotation (1-2 genomes) from PacBio long reads | $1,000 | $1,180 | $1,250 |
| Microbial genome assembly & annotation (3-5 genomes) from PacBio long reads | $1,500 | $1,770 | $1,875 |
| Microbial genome assembly & annotation (6-10 genomes) from PacBio long reads | $2,000 | $2,360 | $2,500 |
| Bacterial draft genome: sequencing using Illumina short reads/assembly/annotation | $1,000 | $1,180 | $1,250 |
| Transcriptome annotation (basic) | $500 | $590 | $625 |
| RNA-Seq or sRNA-Seq analysis (up to 24 samples, single mapping reference) | $1,500 | $1,770 | $1,875 |
| RNA-Seq or sRNA-Seq analysis (25 to 72 samples, single mapping reference) | $2,000 | $2,360 | $2,500 |
| RNA-Seq or sRNA-Seq analysis (above 73 to 96 samples, single mapping reference) | $2,500 | $2,950 | $3,125 |
| RNAseq or sRNAseq analysis (for each additional mapping reference) | $500 | $590 | $625 |
| Single Cell RNA-Seq analysis | $1,000 | $1,180 | $1,250 |
| GO tag and InterProScan annotation | $300 | $354 | $375 |
| SNPs detection/calling/filtering (up to 24 samples) | $1,500 | $1,770 | $1,875 |
| GBS analysis using STACKS (up to 96 samples) | $1,500 | $1,770 | $1,875 |
| Microbiome analysis (up to 24 samples) | $500 | $590 | $625 |
| Microbiome analysis (25-96 samples) | $1,000 | $1,180 | $1,250 |
| Bacterial genome submission to NCBI | $250 | $295 | $313 |
| PacBio: CCS reads generation | $300 | $354 | $375 |
| Hourly rate for custom jobs and/or personnel training | $75 | $89 | $94 |
Please contact GGBC to inquire about pricing for any bioinformatics services not listed above.
