Short-Read Transcriptomics
RNA-Seq (RNA sequencing), also called whole transcriptome shotgun sequencing (WTSS), uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment in time.
RNA-Seq is used to analyze the continuously changing cellular transcriptome. Specifically, RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/SNPs and changes in gene expression over time, or differences in gene expression in
different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, and small RNA, such as miRNA.
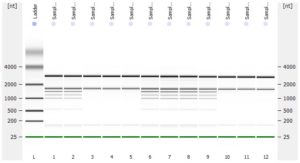
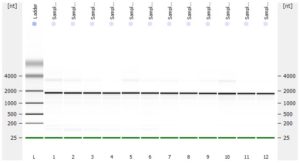
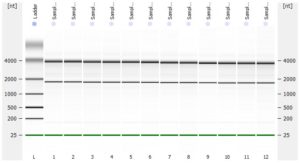
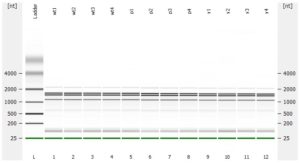
The RNA-Seq workflow should start with experimental design (GGBC consultation). After designing the experiment, a suitable method is chosen to isolate and purify RNA. GGBC accepts purified total RNA as the input to a range of NGS library preparations. For each library preparation, the workflow starts with the quality assessment of the input RNA. The RNA quality is critical for successful RNA-Seq experiment. is critical for maximizing the quality of the returned data.
Consultation and Assistance
For technical questions about existing or new Illumina projects, please contact the following Genomics Professional:
- GGBC (ggbc@uga.edu) for RNA-Seq and sRNA-Seq libraries
More contacts
Please visit our “All Inquiries” page for detailed information about who you should contact at GGBC to receive a quick and accurate response.
Sample Preparation
Libraries prepared with the following sample preparation kits at the GGBC can be used on any of Illumina’s next generation sequencing instruments including MiSeq, NextSeq 500 and HiSeq.
Crucial recommendations for all samples
- Use fluorometric based methods for quantification (Qubit or PicoGreen) of the template DNA instead of UV spec based methods (e.g., NanoDrop) to obtain accurate DNA measurement.
- DNA should have absorbance ratio values of 1.8–2.0.
- RNA samples can be suspended in RNase-free water or 1X TE buffer prepared with RNase-free water.
- RNA integrity should be assessed using the Agilent Bioanalyzer (or any similar system).
- Please add any QC information (Qubit data, Agilent run, fragment analyzer run, gel picture,…) available to the online order form.
- If you are are submitting 24 or more samples, please submit samples in a 96-well plate and include an Excel file with the sample layout in your order.
- PLEASE USE V BOTTOM PLATES AND NOT ROUND/FLAT BOTTOM PLATES.
RNA is a sensitive material. RNA should be kept on dry or wet ice during transportation to the GGBC. Wet ice is recommended only for samples that are being transported locally within Athens. If you are traveling from farther away to submit your RNA samples, keep them on dry ice during transit. The GGBC will not accept RNA samples that are not stored properly during transit.
Transcriptomics Library Prep
| Library Prep Type | Description | Concentration requirement | Volume requirement |
|---|---|---|---|
| Stranded or unstranded RNA | We currently use Kapa Biosystems RNA chemistry for constructing stranded libraries. Non-stranded RNA libraries can be prepared upon request | 4 - 160 ng/uL of total RNA, or 0.4 - 16 ng/uL of rRNA depleted or poly(A)-enriched RNA | ≥ 30 uL |
| Ribo-depleted stranded or unstranded RNA | Treatment of the total RNA to remove cytoplasmic rRNA prior to strand specific or unstranded library preparation. We currently use QIAseq FastSelect Fly, Yeast, Worm, Fish, Bacterial, Plant and HMR kits | >100 ng/uL of total RNA | ≥ 15 uL |
| Small RNA | Primarily targets microRNA and other small RNA to generate cDNA from total RNA or purified small RNA | 0.2 - 400 ng/uL of total RNA, or purified small RNA from 1-10 µg total RNA | ≥ 10 uL |
| SMARTer Pico cDNA Synthesis | The SMARTer Pico PCR cDNA Synthesis Kit provides a PCR-based method for producing high-quality cDNA from picogram quantities of total RNA. | ≥ 20 pg/uL of total RNA | ≥ 55 uL |
| Ready-to-pool libraries/ Ready-to-run pool | ≥20 µl of your ready to run library at a 10 ± 4 nM concentration in 10 mM Tris pH 8. If you woµLd like to use a custom primers for your run, please submit ≥80 µl at a concentration of 10 mM. | ||
RNA-seq Libraries
The GGBC currently prepares mRNA-Seq libraries from poly-adenylated total RNA for the Illumina platform using a strand orientation preserving mRNA-Seq method (stranded). Polyadenylated mRNA is first captured with oligo (dT)n modified paramagnetic beads, washed to remove unbound RNA and eluted into a fragmentation solution. After fragmentation with heat in the presence of metal ions, randomly primed first strand synthesis is initiated. For second strand marking and strand orientation preservation, dUTP is incorporated during the second strand synthesis. End repair, A-tailing and Illumina adapter ligation followed by amplification with a dUTP intolerant polymerase completes the library synthesis.
For non-polyadenylated RNA and for samples where the analysis of non-polyadenylated RNAs is desired, the GGBC uses the QIAseq FastSelect rRNA depletion reagents to remove the structural RNA. The standard RNA-Seq protocol is then initiated at the fragmentation and randomly primed first strand synthesis step.
The small RNA-seq library are prepared using a gel-free method starting with total RNA as input. Truncated Illumina specific adapters are directly ligated to the 3’ OH group and the 5’ phosphate group of the small RNAs using modified RNA ligases, followed by reverse transcription and PCR. The gel-free method is more robust and cost effective than other gel-based methods.